A new framework for trusting modeling predictions in biology
Collaborators
A new approach that combines multiple molecular pathway models allows scientists to better predict how our cells adapt and repair. These insights could help improve muscle performance and recovery.
By Kristy Hamilton
Image generated with Google Gemini.
Inside each of our cells, tiny signaling molecules guide critical decisions: when a cell should grow, divide, or self-destruct. Those decisions can affect muscle repair, long-term tissue adaptation to exercise, and even fatigue responses.
To make sense of this complexity, biologists rely on mathematical models, which themselves can be complex with designs that must account for unknown elements and limited experimental data. The resulting proliferation of models – sometimes dozens to hundreds – each with slightly different assumptions, presents a new quandary: How do you choose which one to use?
“When researchers rely on just one model, their predictions depend entirely on the assumptions built into that model. If the model is missing something or has errors, the predictions can be wrong,” said Padmini Rangamani, a Professor of Pharmacology and Mechanical and Aerospace Engineering and Wu Tsai Human Performance Alliance faculty at UC San Diego.
A study in Nature Communications by Rangamani, Nathaniel Linden-Santangeli, and colleagues suggests a new way. By leveraging the statistical technique Bayesian multimodel inference (MMI), researchers show it’s possible to robustly combine models rather than pick a single “winner.” The approach boosts confidence in predictions about how cells behave.
The Challenge: Too Many Models
Biologists use mathematical models to understand how cells work. But:
- Models have to deal with unknowns and limited data
- There are often dozens or even hundreds of versions for the same pathway
- Picking just one can give biased or wrong results
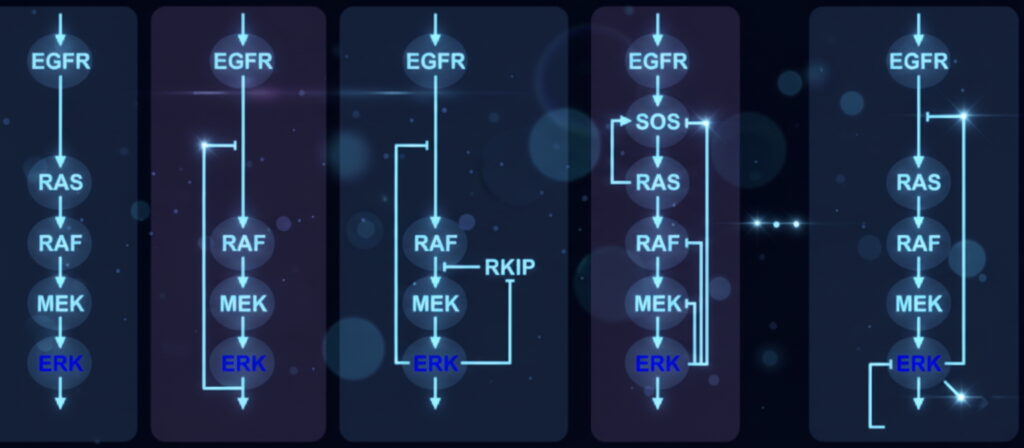
The researchers used models of ERK signalling to apply Bayesian MMI for systems biology. The Bayesian MMI workflow is applicable whenever multiple models are available. Image generated with Google Gemini.
A Smarter Way: Bayesian Multimodel Inference
The team focused on a signaling circuit known as the ERK pathway, a molecular communication chain that controls how cells respond to growth signals and regulate muscle growth, repair, and adaptation. This pathway has been modeled extensively, with more than 125 versions that exist in the BioModels database alone. Each version attempts to capture the pathway’s dynamics, but with different guesses about which steps matter most.
Rather than picking one model, the team applied Bayesian multimodel inference to a set of 10 ERK models. “We chose these 10 models, because they had the same inputs and outputs and because they represented the ERK signaling pathway with a variety of mechanisms,” said Rangamani.
The method assigns weights to each model depending on how well it predicts experimental data, then combines their forecasts. The result: predictions that are more reliable and robust when additional models are swapped in or out.
“While we still have to make modeling assumptions with Bayesian multimodal inference, it allows us to rigorously leverage a set of plausible assumptions and use the data to select those that contribute most,” said Rangamani.
Surprising Findings
In one test, the method helped explain puzzling measurements showing that ERK activity differs depending on its location inside the cell.
“Bayesian multimodel inference helped us to select the set of mechanistic hypotheses that best agreed with the data [we had from past literature],” added Rangamani. “We were most surprised to find two different mechanisms–Rap1 activation and ERK negative feedback–were necessary for the sub-cellular differences in ERK signaling and were able to develop a new model based on these findings.”
Broader Implications
While the study focused on ERK, the implications are far broader. Multimodel inference could be applied to questions ranging from tumor growth to immune signaling – essentially anywhere multiple plausible models coexist.
Still, challenges remain. The methods can be computationally demanding, since each model requires extensive parameter estimation. Some versions of the technique, like full Bayesian model averaging, are accurate but also complex to implement.
Not all Bayesian multimodel inference methods are equal, and choosing the right one depends on the system and data available. In this study:
- Pseudo-Bayesian model averaging” performed best overall. It provided accurate predictions, quantified uncertainty, and is widely applicable.
- “Stacking” tended to favor only the single best-performing model, especially when all models made similar predictions, which can limit its usefulness.
- “Full Bayesian model averaging” can reduce predictive uncertainty but may increase predictive error compared to the best models. It also requires additional computations, making it more computationally demanding.
The researchers found that a compromise approach, called pseudo-Bayesian model averaging, offered the best balance between rigor and practicality. They also found that using multiple of these methods together can increase confidence in model selection.
In the messy world of systems biology, perhaps combining models – rather than betting on a single one – can provide clearer insights.
The work is part of the Wu Tsai Human Performance Alliance Multiscale Athlete Moonshot.
Co-authors include Nathaniel Linden-Santangeli, Jin Zhang, and Boris Kramer.
Latest News

December 10, 2025
Study suggests how eccentric resistance exercises might strengthen tendons

November 21, 2025
Recordings Now Live: Female Athlete Research Meeting 2025
September 23, 2025
In sprinting, where the foot lands may be key to speed
Get Engaged
Join our mailing list to receive the latest information and updates on the Wu Tsai Human Performance Alliance.
